Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
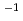 carbon beam
carbon beamTsuda, Shuichi; Sato, Tatsuhiko; Takahashi, Fumiaki; Satoh, Daiki; Endo, Akira; Sasaki, Shinichi*; Namito, Yoshihito*; Iwase, Hiroshi*; Ban, Shuichi*; Takada, Masashi*
Radiation Protection Dosimetry, 143(2-4), p.450 - 454, 2011/02
Times Cited Count:5 Percentile:38.51(Environmental Sciences)A wall-less tissue equivalent proportional counter, wall-less TEPC, has been designed and used for the measurement of the y distributions for energetic heavy ions in order to verify a biological dose calculation model incorporated in the PHITS code. It is found that the dose-mean value of y obtained by the wall-less TEPC is 50 - 60% of the LET of the argon ions in water, since the delta-rays with relatively low y can be measured.
Sato, Tatsuhiko; Watanabe, Ritsuko; Kase, Yuki*; Tsuruoka, Chizuru*; Suzuki, Masao*; Furusawa, Yoshiya*; Niita, Koji*
Radiation Protection Dosimetry, 143(2-4), p.491 - 496, 2011/02
Times Cited Count:33 Percentile:91.34(Environmental Sciences)We reanalyzed the survival fraction data, using the microdosimetric-kinetic (MK) model implemented in the PHITS code. It is found from the analysis that the MK model successfully accounts for the cell survival-fractions under a variety of irradiation conditions, using only y* parameter.
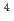 He
He ion irradiation
ion irradiationYokoya, Akinari; Shikazono, Naoya; Fujii, Kentaro; Noguchi, Miho; Urushibara, Ayumi
Radiation Protection Dosimetry, 143(2-4), p.219 - 225, 2011/02
Times Cited Count:3 Percentile:25.9(Environmental Sciences)Multiple single-strand breaks (m-SSBs), which are predicted to be preferentially induced by high LET radiation, would be underestimated if one uses the conventional method using plasmid DNA, because m-SSBs will not cause additionally conformational changes if they are on the same or on the opposite strand but separated each other sufficiently so as not to induce a double strand break. In order to observe the invisible m-SSBs, we have developed a novel technique using DNA denaturation. The m-SSBs arising in both strands of DNA are revealed as molecular size change in single strand DNA (SS-DNA) by gel electrophoresis. We have applied this method to the X- and He ion irradiated sample of hydrated pUC18 plasmid DNA. A half of SS-DNA population remains as intact within the experimental resolution (
ion irradiated sample of hydrated pUC18 plasmid DNA. A half of SS-DNA population remains as intact within the experimental resolution (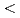 140 bases) for both irradiations. Contrary to our initial expectation, these results indicate that SSBs are not multiply induced over 140 bp even by high-LET irradiation.
140 bases) for both irradiations. Contrary to our initial expectation, these results indicate that SSBs are not multiply induced over 140 bp even by high-LET irradiation.
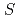 -values in a mouse voxel phantom
-values in a mouse voxel phantomMohammadi, A.; Kinase, Sakae
Radiation Protection Dosimetry, 143(2-4), p.258 - 263, 2011/02
Times Cited Count:17 Percentile:77.31(Environmental Sciences)Watanabe, Ritsuko; Wada, Seiichi*; Funayama, Tomoo; Kobayashi, Yasuhiko; Saito, Kimiaki; Furusawa, Yoshiya*
Radiation Protection Dosimetry, 143(2-4), p.186 - 190, 2011/02
Times Cited Count:16 Percentile:75.64(Environmental Sciences)Microscopic energy deposition pattern in an ion track is thought to affect on the spatial distribution of DNA damage as well as the damage spectrum. In this study, we focus on the intra-track spatial distribution of DNA damage in cellular condition based on the energy deposition pattern for each ion obtained by the detailed Monte Carlo track structure simulation. The estimation was performed for C and Ne ions with similar LET around 440 keV/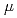 m. As a result, radial DNA damage distribution shows different pattern for C and Ne ions. That is, DSBs or non-DSB type clustered damage are formed in the limited central area while the isolated damages as SSBs and base lesions are spread in larger area. Such tendency is more clearly shown for Ne ions than C ions. This result shows good agreement with the previously obtained experimental observation at TIARA, which indicates the different types of DNA damage shows different distribution pattern around C and Ne projectiles in cell nuclei.
m. As a result, radial DNA damage distribution shows different pattern for C and Ne ions. That is, DSBs or non-DSB type clustered damage are formed in the limited central area while the isolated damages as SSBs and base lesions are spread in larger area. Such tendency is more clearly shown for Ne ions than C ions. This result shows good agreement with the previously obtained experimental observation at TIARA, which indicates the different types of DNA damage shows different distribution pattern around C and Ne projectiles in cell nuclei.
Ouchi, Noriyuki
Radiation Protection Dosimetry, 143(2-4), p.365 - 369, 2010/12
Times Cited Count:1 Percentile:10.7(Environmental Sciences)Recent years, risk assessments of radiation induced cancer are getting worth for the public health view points. In general, we know that carcinogenesis by low dose radiation will start from DNA damage by ionizing radiation. After the long time period, these very small effects will appear on a cellular scale by accumulation of various intracellular biological responses and finally grow to the tumor with clonal expansion of cancer cell. Thus, the biological radiation effects are phenomena with a very wide scale from DNA damage to the tumor, so the risk estimation of low dose radiation is difficult to study by the experiments. To overcome these difficult situations at low dose radiation effects problem, it is good to study process of carcinogenesis using biologically based mathematical model. In this presentation, we will introduce our cellular scale mathematical model of tumorigenesis and show some results of statistical calculations about tumor growth curve.
Shikazono, Naoya; Yokoya, Akinari; Urushibara, Ayumi; Noguchi, Miho; Fujii, Kentaro
no journal, ,
To evaluate the spatial distribution of radiation-induced strand breaks, we have developed a simple analytical model based on several assumptions for the generation of strand breaks in plasmid DNA after irradiation. The model assumes that (1) a radiation track hits a plasmid with a probability following a Poisson distribution, (2) the hit randomly generates strand break(s) with its number following a Poisson distribution within a single, relatively short DNA segment, and (3) a double strand break is formed when each strand has at least one strand break in the segment hit by the track. To find out whether the model is valid, we compared the calculated values to experimental data obtained by the plasmid DNA assay. The model described well the experimental results of hydrated plasmids exposed to radiation with low linear energy transfer, suggesting that the model could be useful in estimating the extent of damage clustering.
Noguchi, Miho; Urushibara, Ayumi; Yokoya, Akinari; O'Neill, P.*; Shikazono, Naoya
no journal, ,
It is proposed that a single track of ionizing radiation induces clustered DNA damage sites and that their complexity increases with increasing LET. Non-DSB clustered damage is thought to contribute to the biological effects of radiation such as mutation. In this study, we have investigated the mutagenicity of clustered DNA damage containing SSB and base damage. We have used a plasmid based assay with Escherichia coli to measure the mutation frequency induced by bistranded clustered damage. As a model of clustered damage, we have synthesized oligonucleotides containing a SSB and/or 8-oxo-7,8-dihydroguanines (8-oxoGs) within the recognition site of the restriction enzyme (Alw26I). Plasmid constructs containing damaged DNA was transfected into wild-type or glycosylase-deficient strains and propagated in cells. The mutation frequency was assessed by the inability of Alw26I to cut the oligonucleotide sequence. The mutation frequency of bistranded clusters containing an 8-oxoG opposite to a second 8-oxoG 2bp apart was the highest of all the types of clusters tested in the present study. When a SSB was included in clusters containing bistranded 8-oxoGs, the mutation frequency is lower in all E. coli strains tested. These results suggest that a SSB located on the same strand to one of the 8-oxoG reduces the mutagenic potential of 8-oxoG. Our studies demonstrate that the mutagenic potential of clusters containing 8-oxoG is modified if a SSB is present within the cluster.